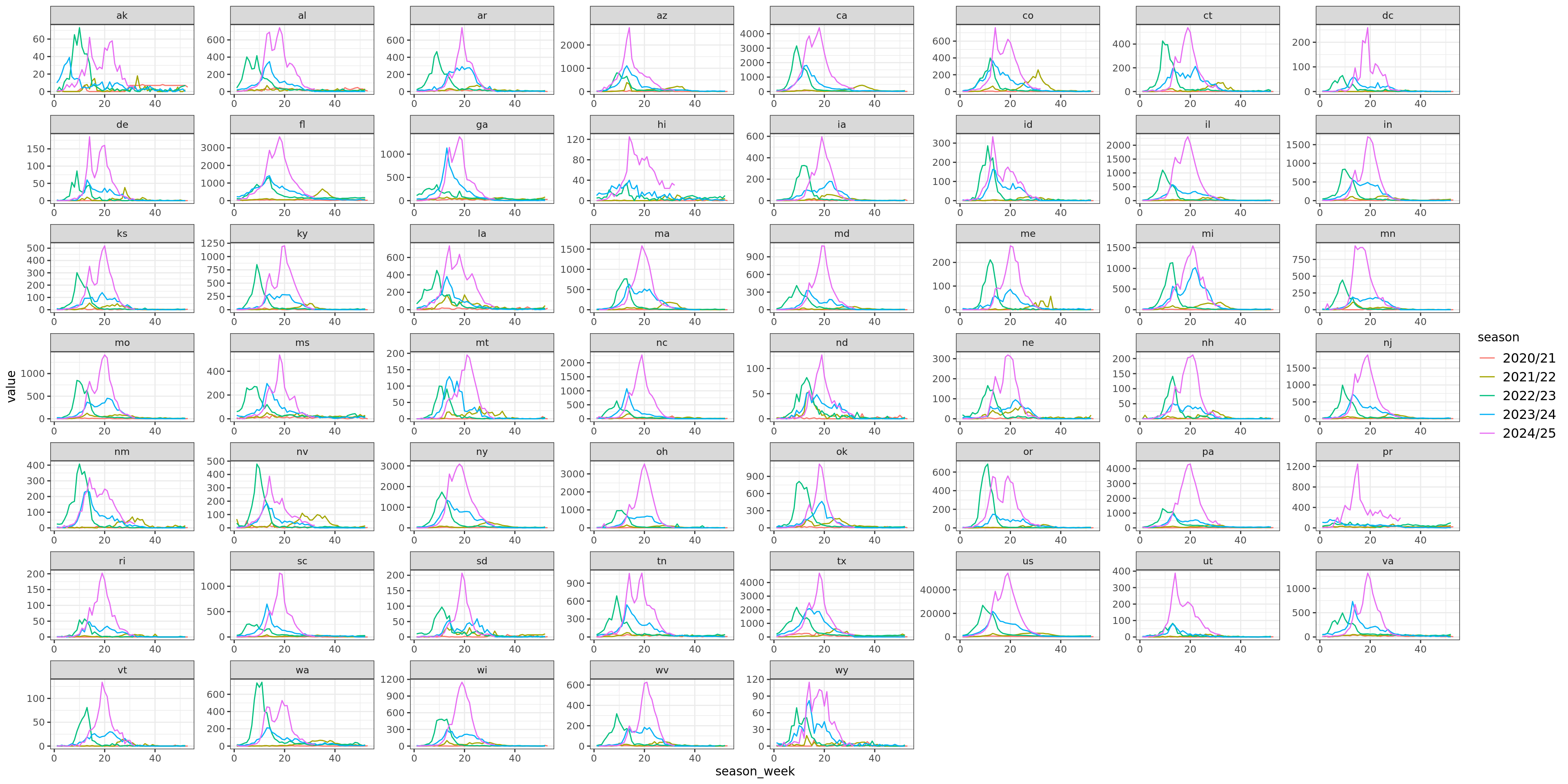
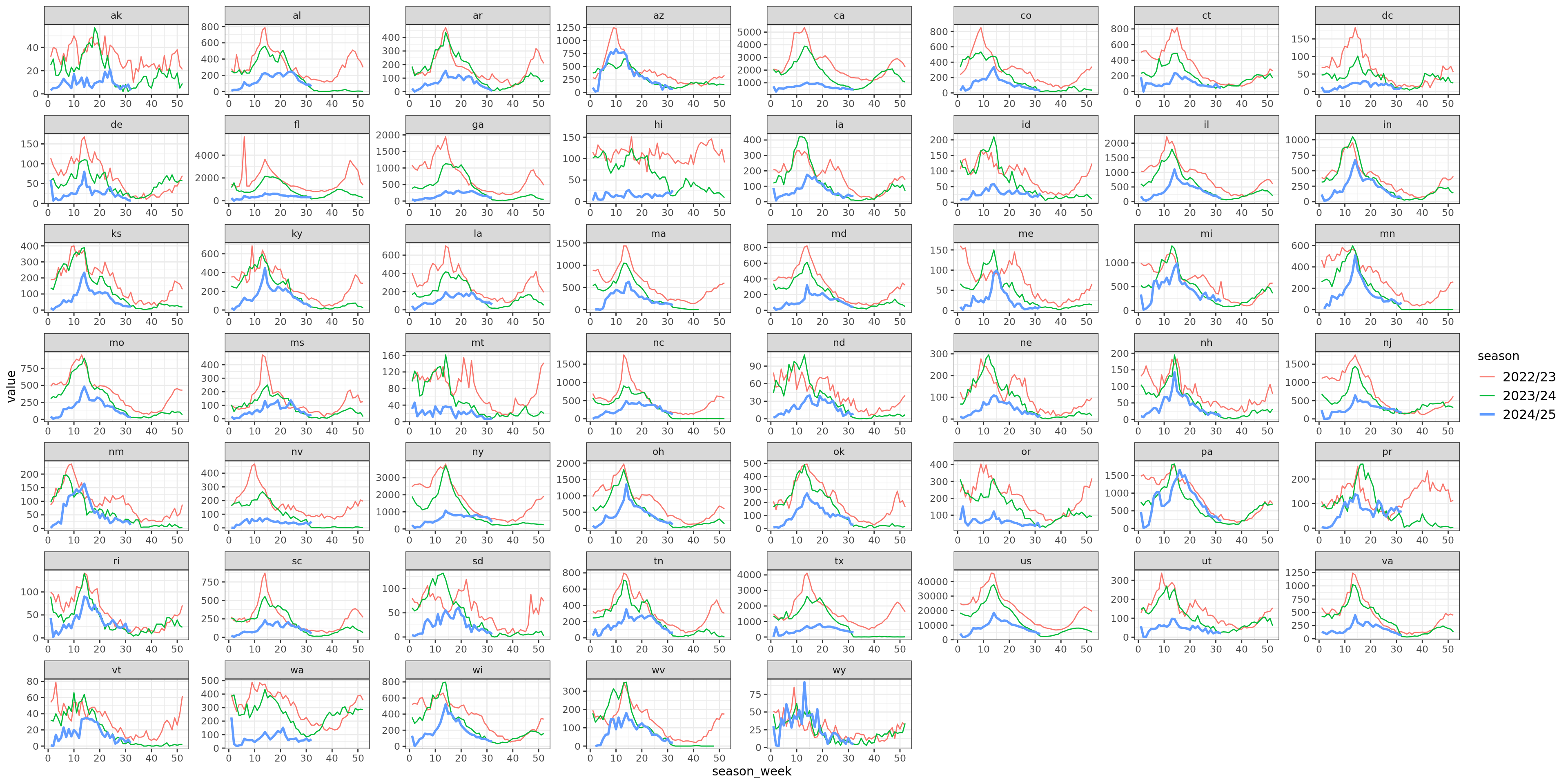
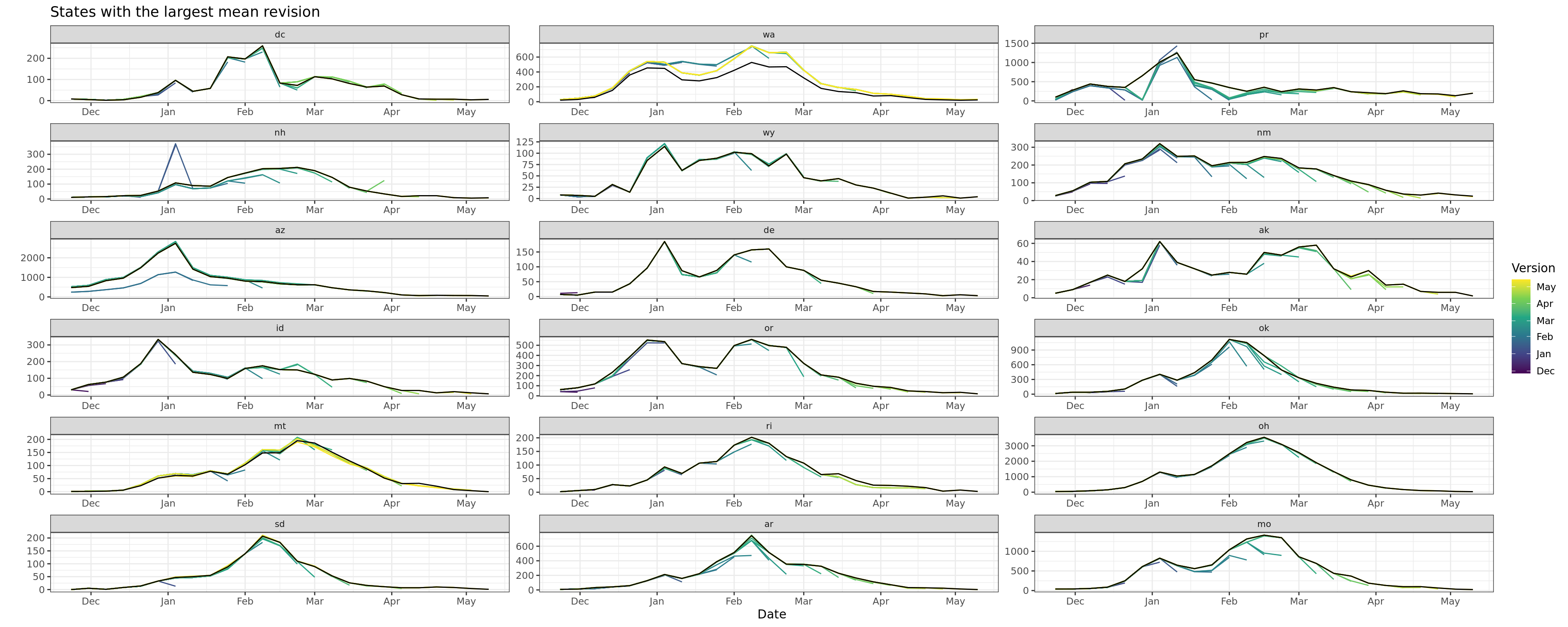
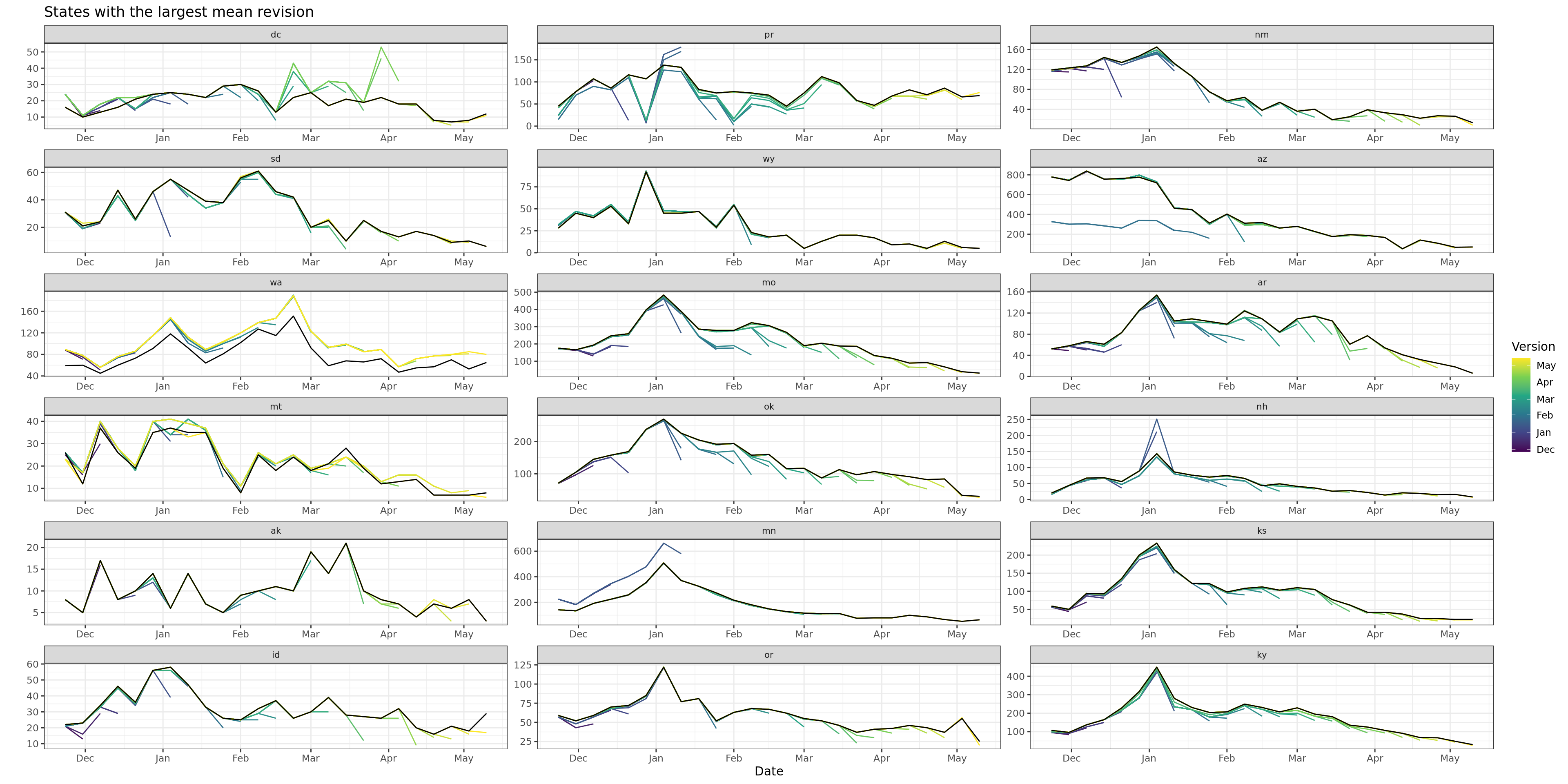
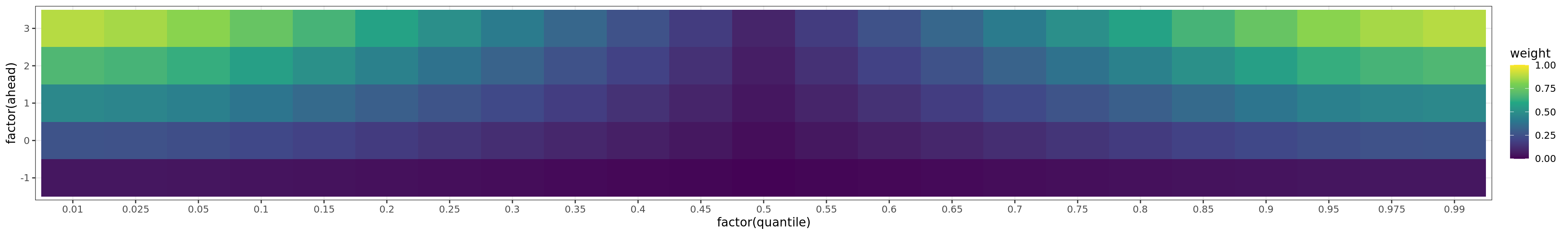
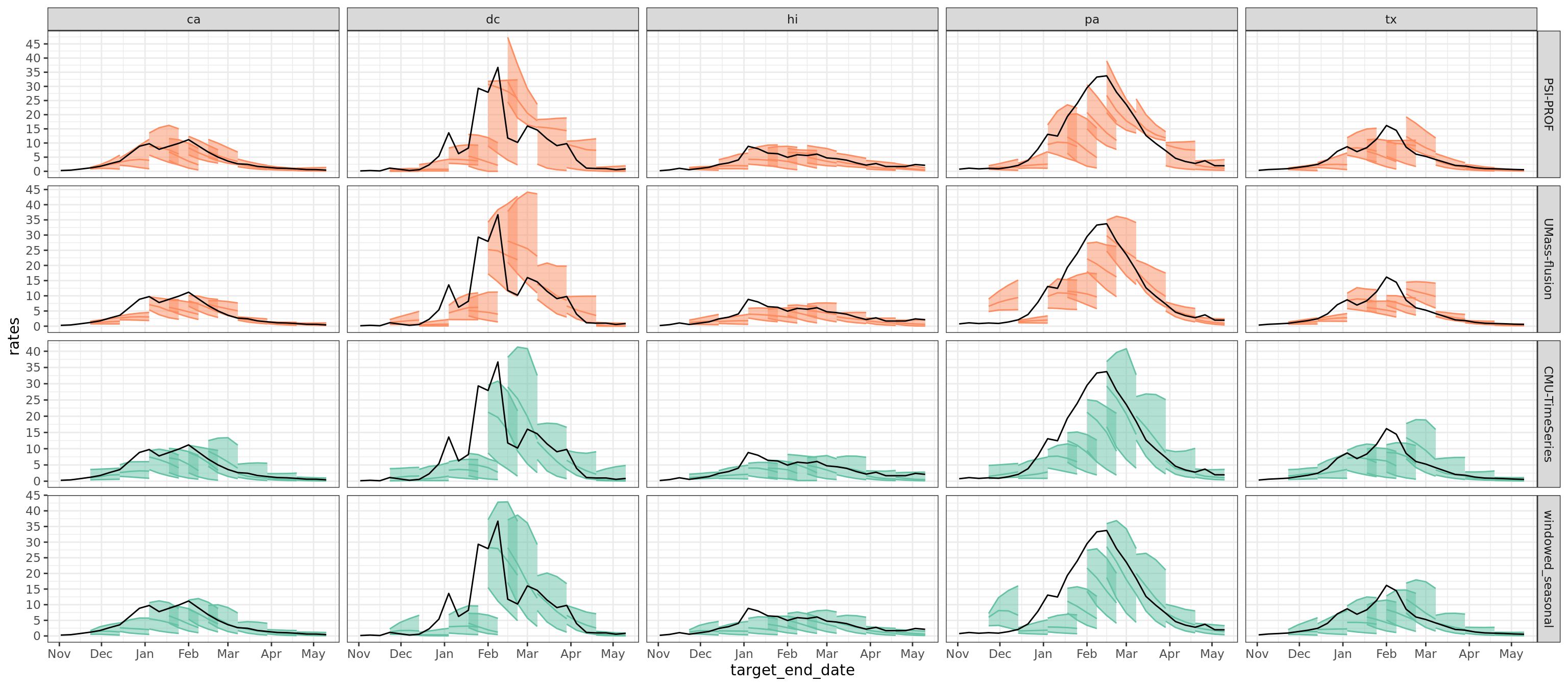
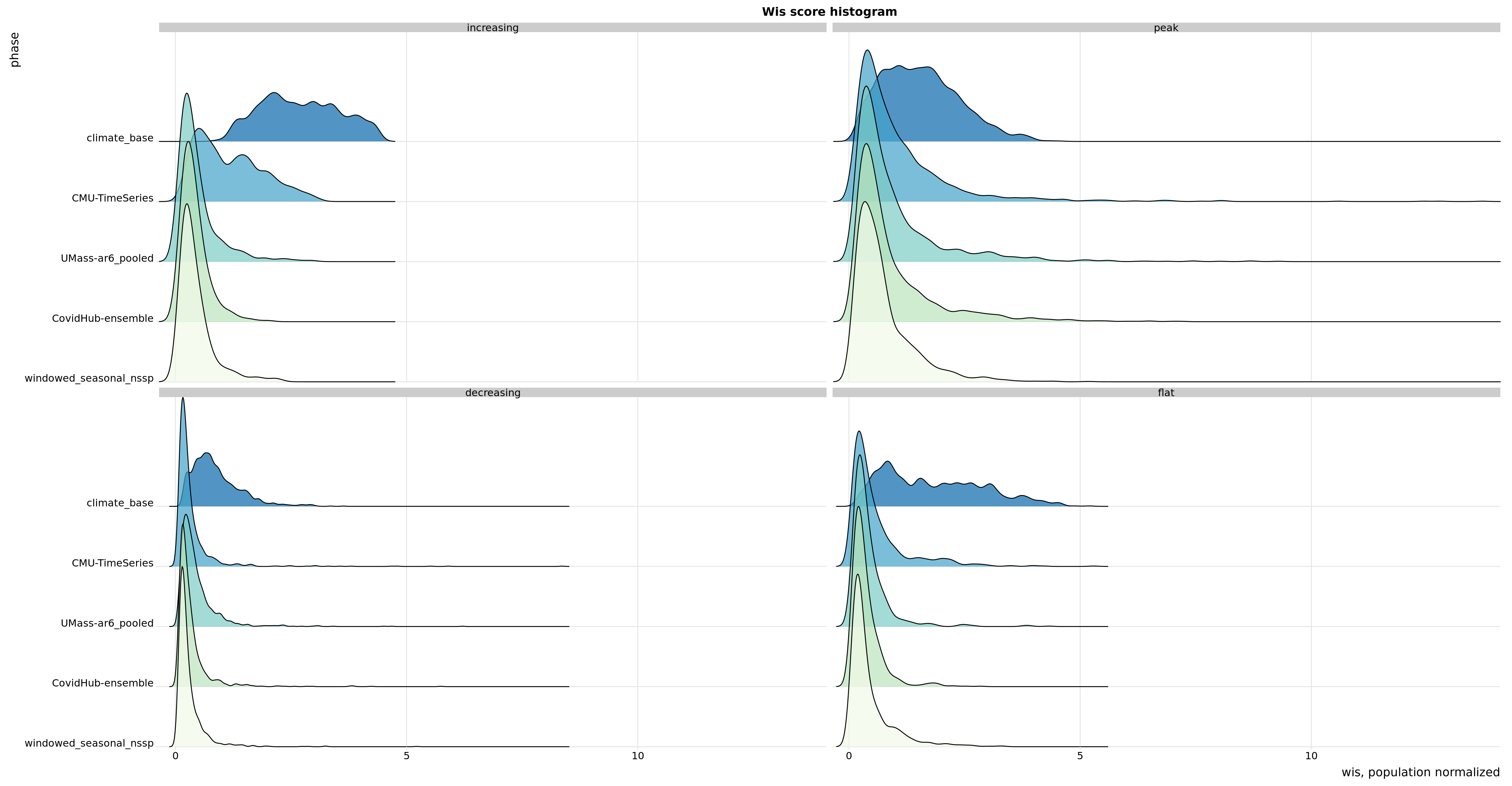
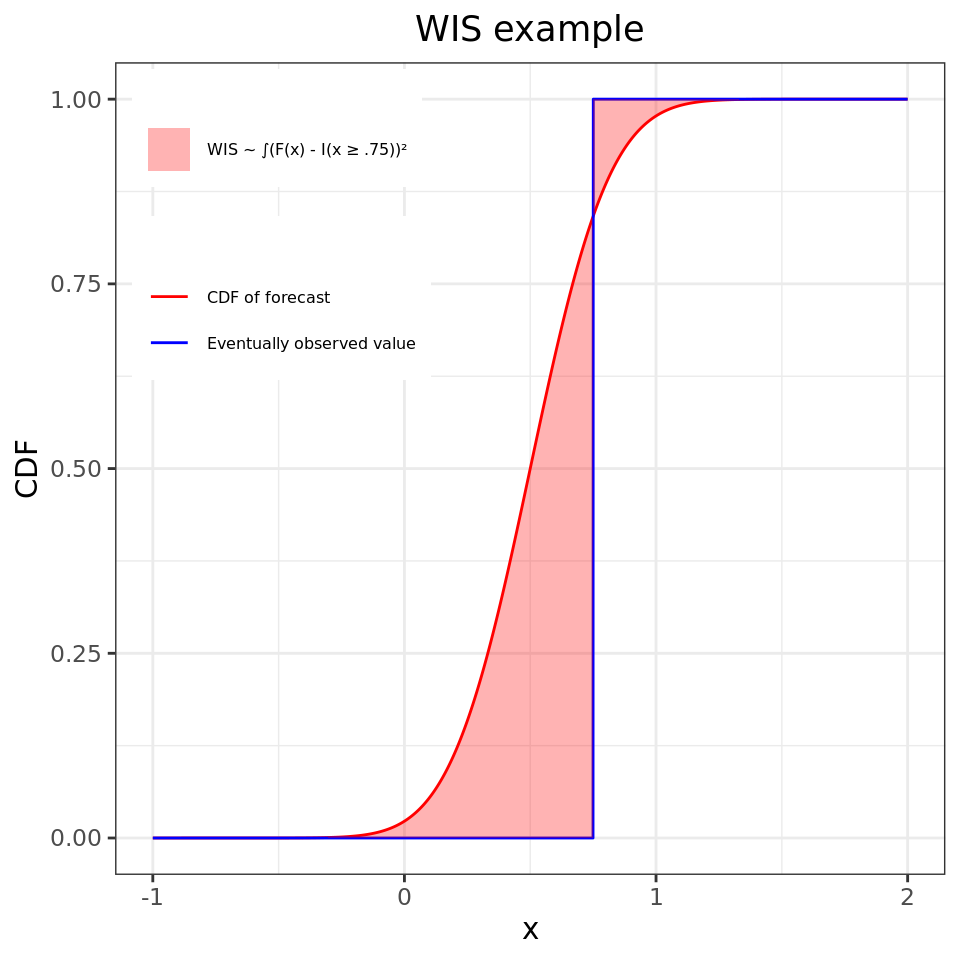
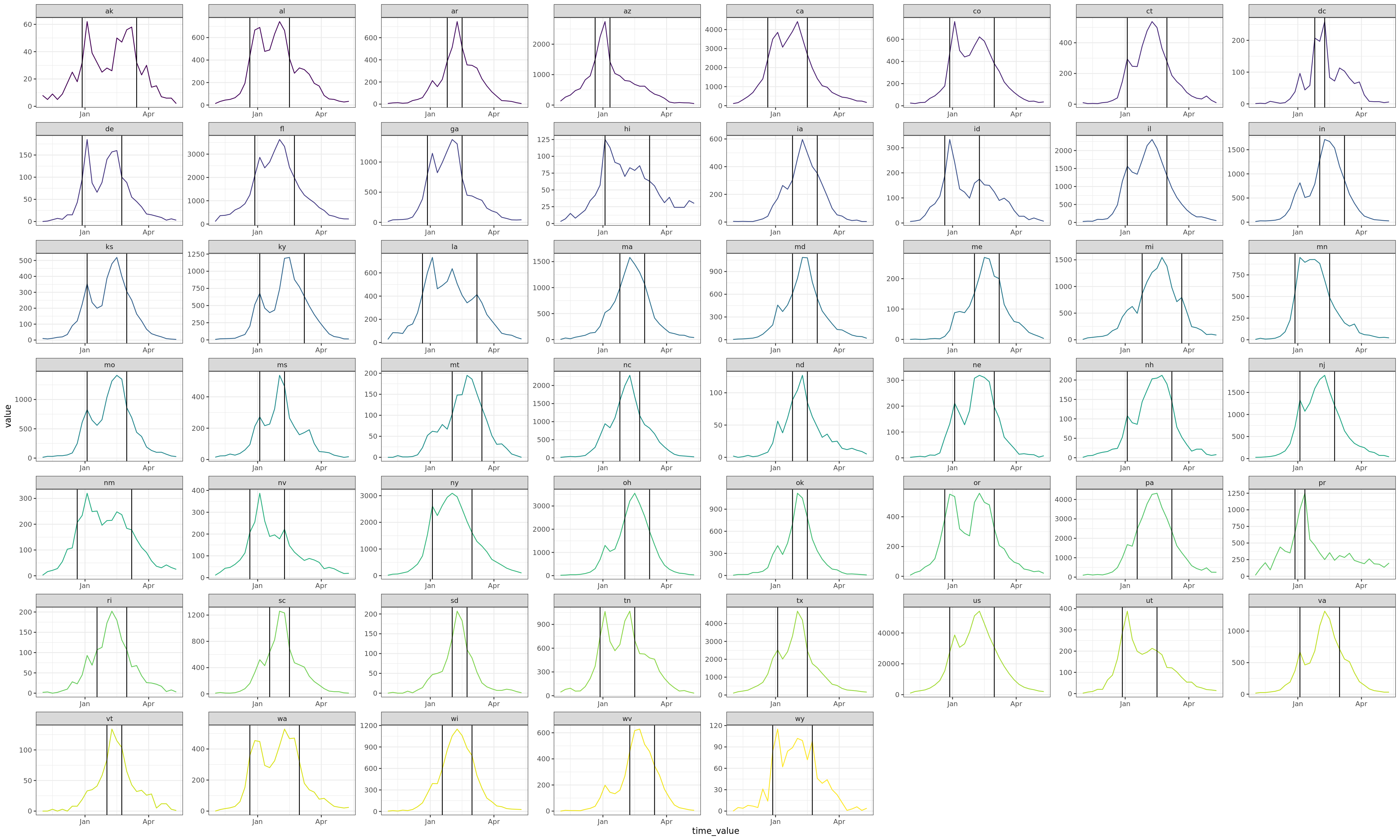
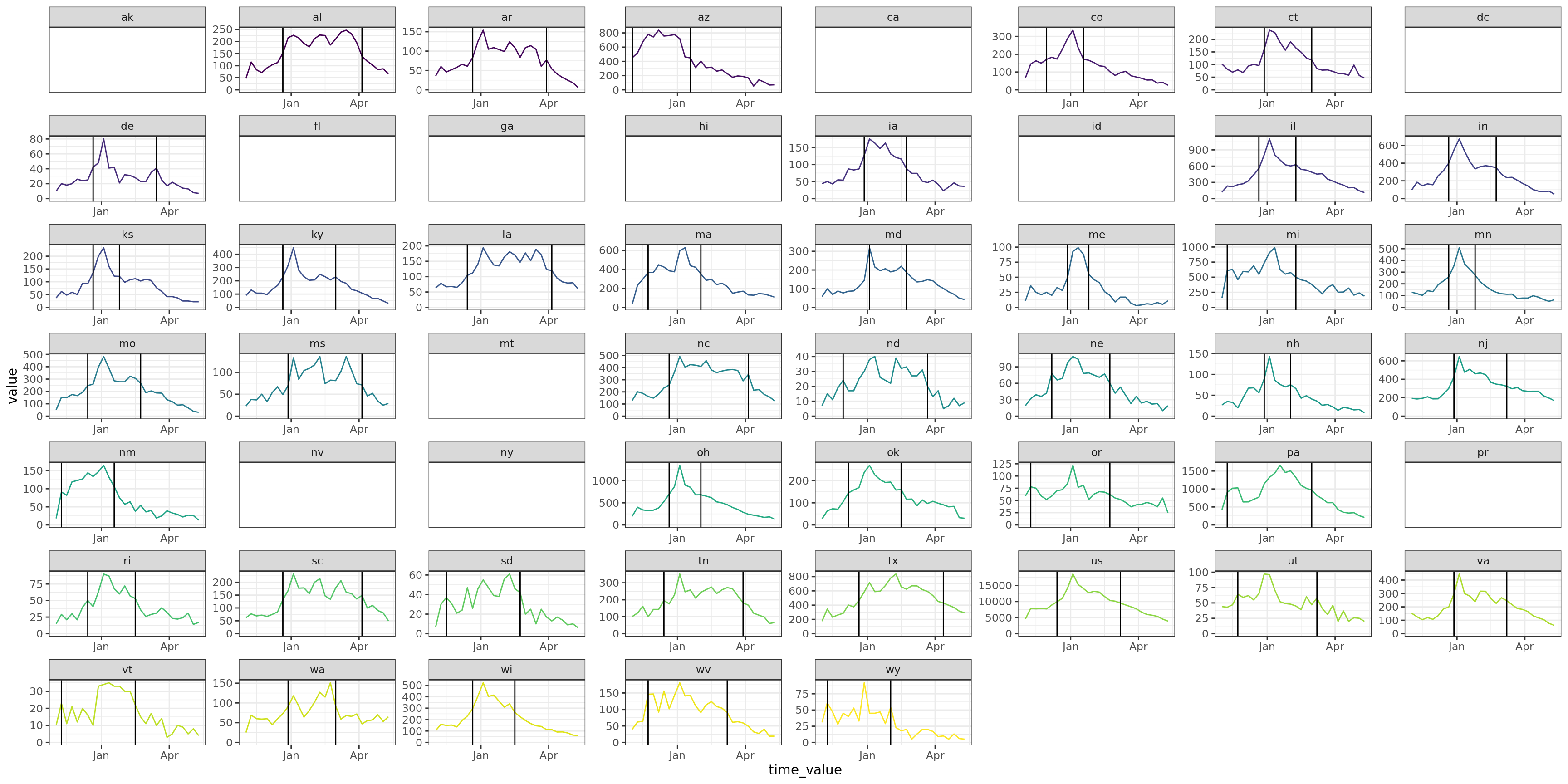
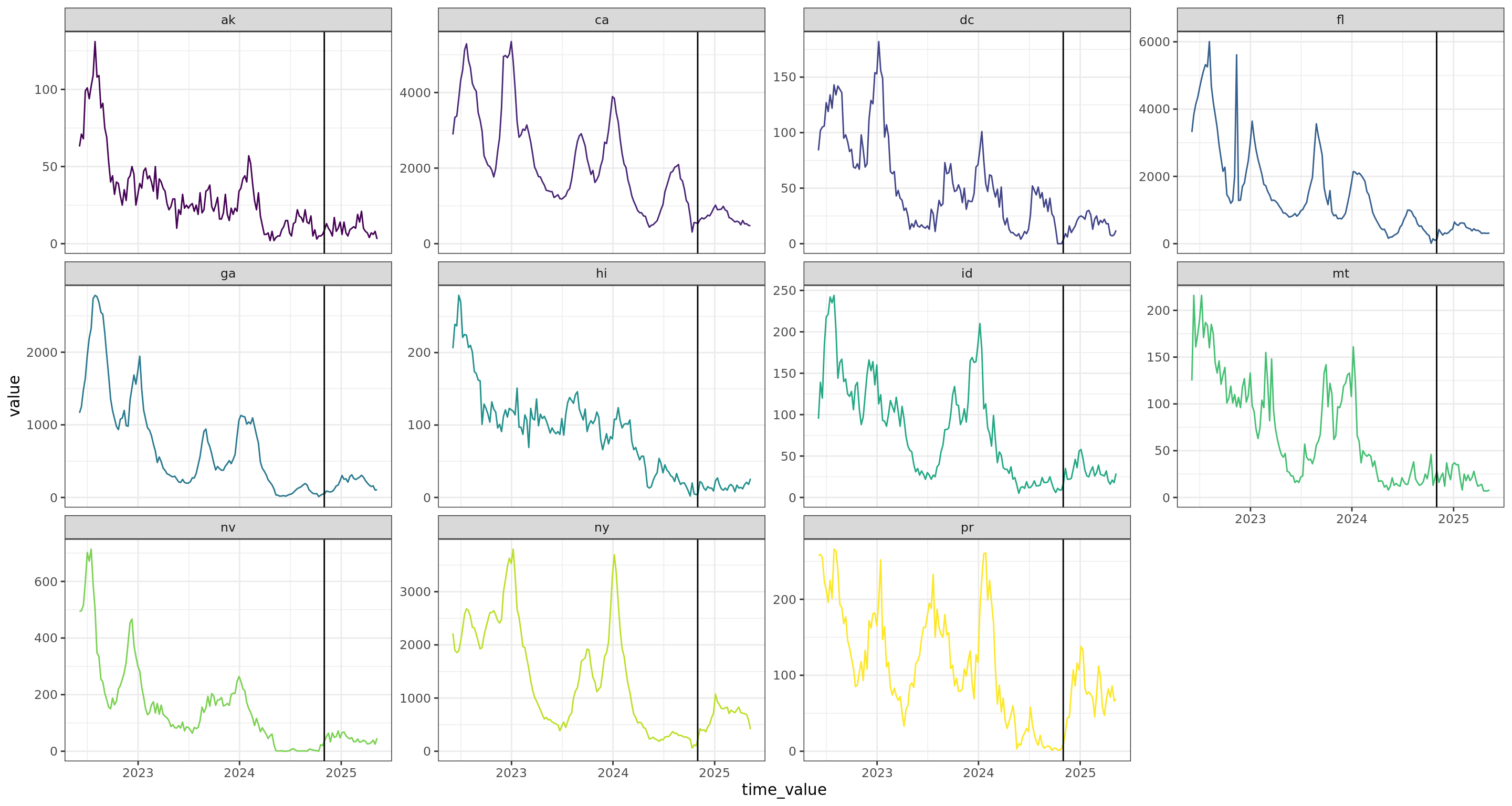
Flu Scores: WIS Rate Histogram
These scores are rates rather than counts, and color gives population. Note there’s an implied fat tail in the distribution of scores. Overall, the distributions of scores are very similar.
In the increasing phase, CMU-TimeSeries has a smaller left tail, but a more concentrated middle, and smaller right tail, so this doesn’t show up in the aggregate. In the peak phase, CMU-TimeSeries has a thinner left tail (meaning we have fewer cases where we’re dead on), which is enough to show up in the aggregate. The decreasing phase is about the same for the compared models.
ggplot(data, aes(x = wis, y = forecaster, fill = forecaster)) + geom_density_ridges(scale = 3, alpha = 0.7) + scale_y_discrete(expand = c(0, 0)) + # will generally have to set the expand option scale_x_continuous(expand = c(0, 0)) + # for both axes to remove unneeded padding scale_fill_brewer(palette = 4) + coord_cartesian(clip = “off”) + # to avoid clipping of the very top of the top ridgeline theme_ridges() + facet_wrap(~phase) + labs(title = “Wis score histogram”) + ylab(“phase”) + xlab(“wis, population normalized”) + theme(plot.title = element_text(hjust = 0.5), legend.position = “none”) + theme(legend.position = “none”)
## Flu Scores: Phase
::: {.notes}
We classify the season into three phases: increasing, peak, and decreasing.
Increasing is before the first time the value exceeds a threshold.
Decreasing is the last time the value dips below the threshold.
Peak is between these two.
The threshold is chosen at 50% of the max value.
Note that our WIS/AE is in the top 5, but everyone's score is quite similar and washed out.
We performed well in the increasing/decreasing phases, but most of our error came in the peak phase.
:::
::: {.cell layout-align="center"}
```{.r .cell-code}
phase_scores <-
flu_scores %>%
filter(forecaster %nin% c("seasonal_nssp_latest")) %>%
left_join(flu_within_max, by = "geo_value") %>%
mutate(phase = classify_phase(target_end_date, first_above, last_above, rel_duration, flu_flat_threshold)) %>%
group_by(forecaster, phase) %>%
summarize(
wis_sd = round(sd(wis, na.rm = TRUE), 2),
ae_sd = round(sd(ae_median, na.rm = TRUE), 2),
across(c(wis, ae_median, interval_coverage_90), \(x) round(Mean(x), 2)),
n = n(),
our_forecaster = first(our_forecaster),
.groups = "drop"
)
sketch <- htmltools::withTags(table(
class = 'display',
style = "font-size: 11px;",
thead(
tr(
th(colspan = 2, "forecaster"),
th(colspan = 2, 'increasing'),
th(colspan = 2, 'peak'),
th(colspan = 2, 'decreasing')
),
tr(
list(th("peak_wis_rank"), th("name"),
lapply(rep(c('wis', 'sd'), 3), th)
)
)
)
))
phase_scores_wider <- phase_scores %>%
select(forecaster, phase, wis, wis_sd, our_forecaster) %>%
pivot_wider(names_from = phase, values_from = c(wis, wis_sd))
wis_max <- phase_scores_wider %>% select(wis_increasing, wis_peak, wis_decreasing) %>% max()
phase_scores_wider %>%
select(forecaster, wis_increasing, wis_sd_increasing, wis_peak, wis_sd_peak, wis_decreasing, wis_sd_decreasing) %>%
arrange(wis_peak) %>%
datatable(
fillContainer = FALSE,
options = list(
initComplete = htmlwidgets::JS(
"function(settings, json) {",
paste0("$(this.api().table().container()).css({'font-size': '", "11pt", "'});"),
"}"),
pageLength = 25
),
container = sketch
) %>%
formatStyle("forecaster", target = c("cell"),
fontWeight = styleEqual(repo_forecasters, rep("900", length(repo_forecasters))),
textDecoration = styleEqual("CMU-TimeSeries", "underline")) %>%
formatStyle(
"wis_increasing",
background = styleColorBar(c(0, wis_max), 'lightblue'),
backgroundSize = '98% 88%',
backgroundRepeat = 'no-repeat',
backgroundPosition = 'center'
) %>%
formatStyle(
"wis_peak",
background = styleColorBar(c(0, wis_max), 'lightblue'),
backgroundSize = '98% 88%',
backgroundRepeat = 'no-repeat',
backgroundPosition = 'center'
) %>%
formatStyle(
"wis_decreasing",
background = styleColorBar(c(0, wis_max), 'lightblue'),
backgroundSize = '98% 88%',
backgroundRepeat = 'no-repeat',
backgroundPosition = 'center'
)
:::